Overview
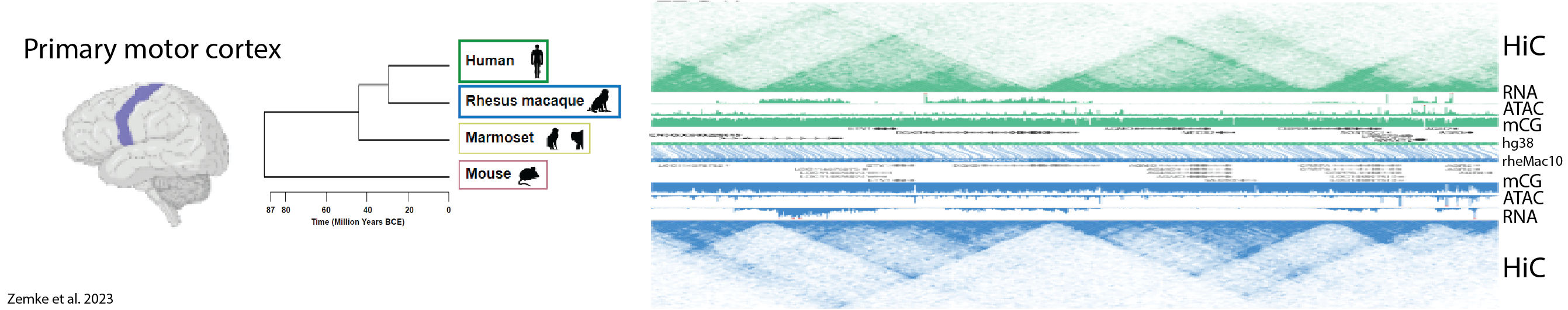
M1 tissue was obtained from three human, macaque, marmoset (Callithrix jacchus) donors, and MOp tissue from eight P56 C57BL/6J male mice (Mus musculus). Mouse MOp was dissected into four subregions (2C, 3C, 4B, 5D) as described in Li et al. 2021 (Nature 2021). Each subregion was pooled from four mice for each replicate, and a total of two replicates was performed for each subregion. Additional experimental details are reported in Zemke et al. 2023.
10X Multiome
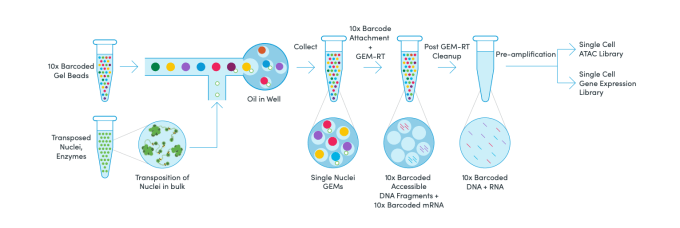
Using 10x multiome (10x Genomics) we simultaneously profiled both transcriptomes and chromatin accessibility. In total, we profiled 40,937 human nuclei, 34,773 macaque nuclei, 34,310 marmoset nuclei, and 47,404 mouse nuclei with 10X Multiome. 10X Genomics.
snm3C-seq
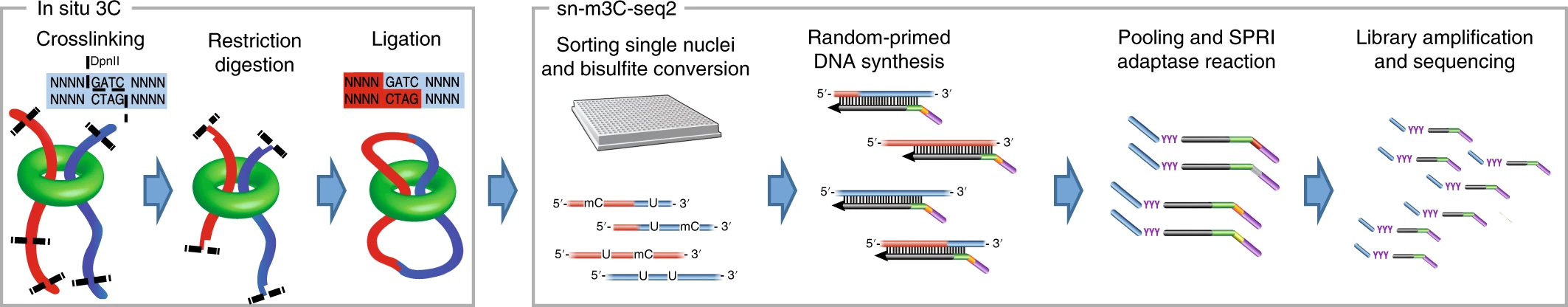
Using snm3C-seq (single-cell Methyl-HiC) we profiled DNA methylation with 3D genome contacts. In total, we profiled 8,198 human nuclei, 5,737 macaque nuclei, 4,999 marmoset nuclei, and 5,349 mouse nuclei with snm3C-seq Lee et al. 2019.
Cell type annotation
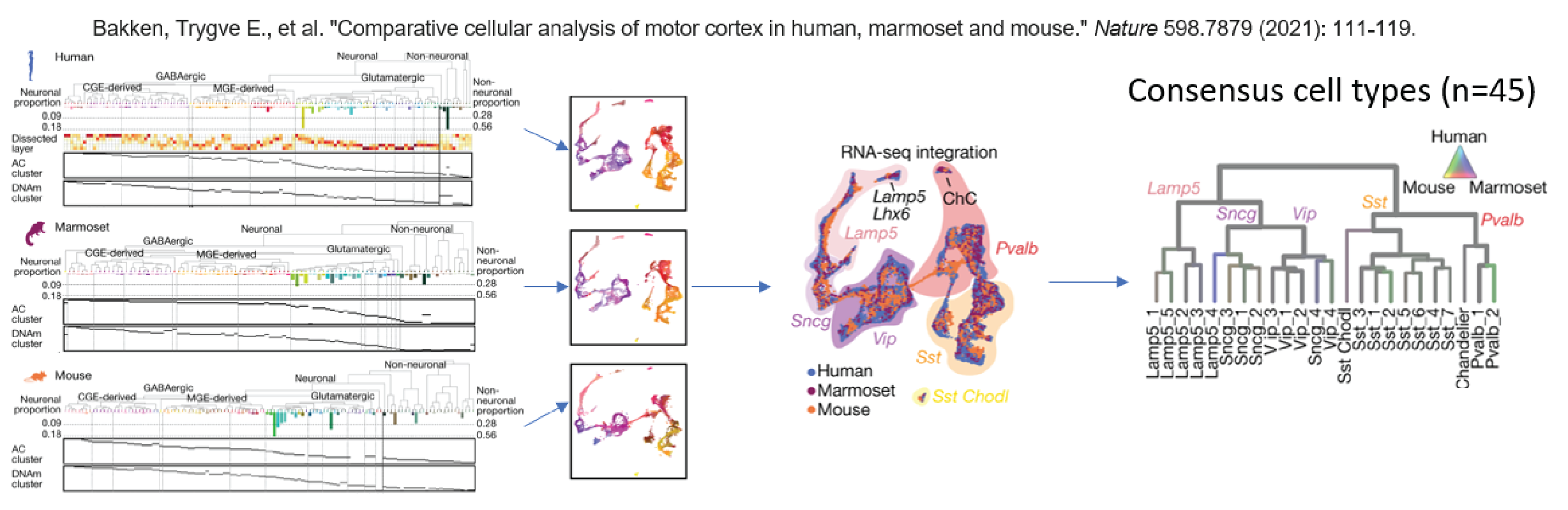
Consensus cell types (n=45), common across species, defined in Bakken et al. 2021 were assigned to both the 10X Multiome and snm3C-seq nuclei from all four species.
Data availability
We are providing data starter kits on Amazon AWS S3 at s3://biccn-challenge in .h5ad or modality specific formats. Request for additional file formats for each modality will be considered!
Using the AWS CLI you can easy download all data files (~130GB) with the following command:
aws s3 sync s3://biccn-challenge . --no-sign-request
For each of the four species you will find:
- An
.h5adfile containing the gene counts from the 10X Multiome.obs.sample: An identifier for donor and sequencing batch.obs.species: Which species the cell came from....: Additional 10X multiome QC metrics....: Cell type annotations from Bakken et al. 2022.
- An
.h5adfile containing the ATAC-seq peaks from the 10X Multiome.obs.Sample: An identifier for donor and sequencing batch....: Addtional ATAC-seq specific QC metrics....: Cell type annotations from Bakken et al. 2022.
- Fragment files from the ATAC-seq componenet of the 10X Multiome experiments.
- These fragment files may contain low-quality cells. Barcodes should be filtered using the corresponding
Species_atac.h5ad.
- These fragment files may contain low-quality cells. Barcodes should be filtered using the corresponding
.h5adfiles containing the gene fractions of mCG, mCH methylation for each cell from the snm3C....: Cell type annotations from Bakken et al. 2022.
- Psuedo-Bulk HiC loops called for each subclass annotation.
Please feel free to reach out with any additional questions or data requests.
Genome Assemblies and Annotations:
- Homo sapiens (Human): GRCh38 (hg38), annotation: hg38 Gencode v33
- Mus musculus (Mouse): GRCm38 (mm10), annotation: mm10 Gencode vM22
- Macaca mulatta (Rhesus monkey): Mmul_10 (rheMac10), annotation: Ensembl release 104
- Callithrix jacchus (white-tufted-ear marmoset): cj1700_1.1 (calJac4), annotation: GCA_009663435.2